The COVID-19 coronavirus SARS-CoV-2 has a telltale “fingerprint” that means it is highly likely to have been made in a lab, new research has found.
Mathematical biologist Dr. Alex Washburne and colleagues Valentin Brutte and Antonius VanDongen have published their research in a preprint that found “a high likelihood that SARS-CoV-2 may have originated as an infectious clone assembled in vitro”.
They explain that this evidence, which they term an “endonuclease fingerprint”, is independent of the evidence relating to the Furin Cleavage Site, which others have suggested is a ‘smoking gun’ for a lab origin.
Dr. Washburne stresses that he is not alleging malign intent or even gain-of-function work in this paper. He writes: “We find no evidence of SARS-CoV-2 being a bioweapon (on the contrary, this looks like an accident) or any gain of function work. We find evidence suggesting SARS-CoV-2 may have been synthesised in the lab with known methods, probably for normal pre-Covid research purposes.”
Professor Francois Balloux has given the study his imprimatur on Twitter, writing: “This is an important piece of work. To me, it looks solid both conceptually and methodologically. I was given advance warning and was able to replicate the key findings. To the best of my knowledge, I confirm the reported patterns are genuine.”
To accompany the study Dr. Washburne has published a Substack and a Twitter thread. The Twitter thread is reproduced in full below – it can be a bit technical in places, but the message it’s conveying should come through clearly enough.
The origin of SARS-CoV-2 is unknown. Some hypothesised two spillover events at the wet market, but methodological flaws make that work inconclusive. We need to know the true origin of SARS-CoV-2 to prevent pandemics.
We examined whether SARS-CoV-2 was synthesised in a lab. We studied a common method for synthesising coronaviruses (CoVs) in the lab. This method was thought to not leave a fingerprint. We found the fingerprint. That fingerprint is in the SARS-CoV-2 genome.
Here’s how you make a CoV in the lab. To make a 30,000 base (30kb) RNA virus in the lab, you need a 30kb DNA clone. To assemble a 30kb DNA clone, scientists glue together several smaller fragments.
A popular method for DNA assembly is ‘golden gate assembly’.
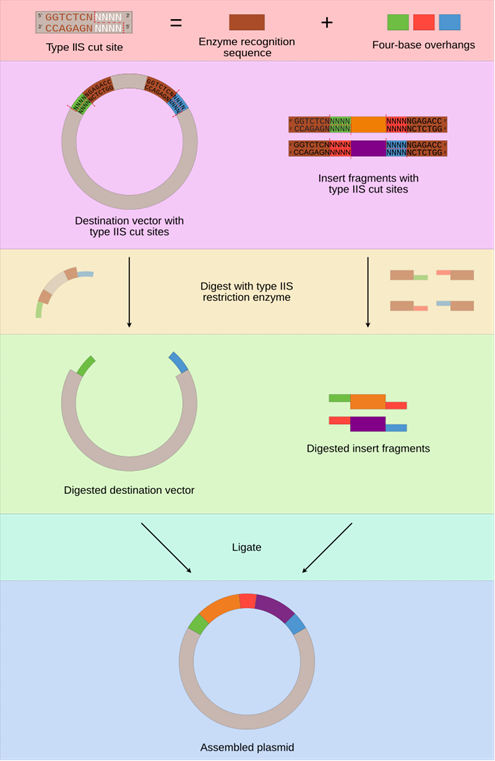
Golden gate assembly requires the DNA sequence have special ‘cutting’ sites (type IIS restriction sites). Cutting sites create three to four nucleotide (nt) ‘sticky ends’. Sticky ends help you ‘paste’ DNA segments together, ensuring faithful assembly of your 30kb DNA copy of a viral genome.
RNA viruses like CoVs are not under selection specifically for this kind of cutting and pasting. So, wild viruses tend to have cutting and pasting sites randomly scattered in their genome. Researchers building viruses in a lab will often add or remove cutting sites.
We collected examples of CoV infectious clones assembled with these type IIS cutting and pasting systems from 2000-2019. We found a clear pattern in how researchers tended to add or remove cutting and pasting sites.
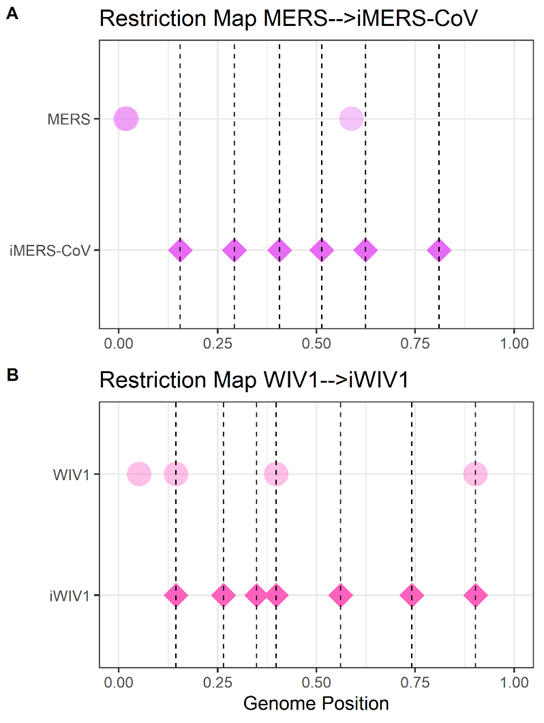
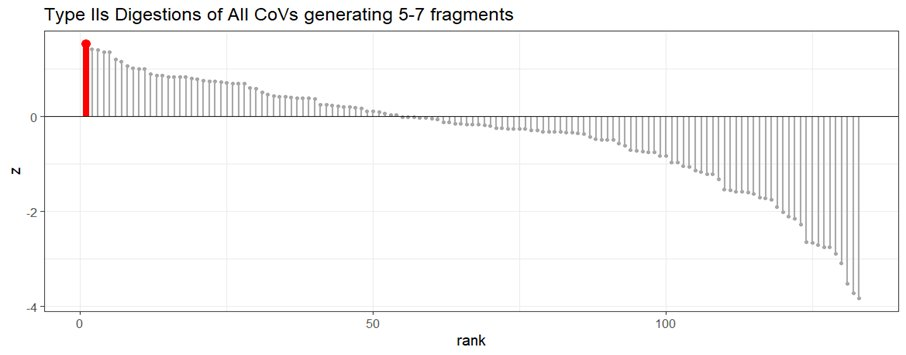
Researchers tend to turn randomly-spaced restriction maps into regularly-spaced ones (A-B). Regular spacing comes from desiring fewer fragments (typically five to eight) while keeping the longest fragment lengths low.
Digesting 70 CoVs with 200-plus restriction enzymes yields a ‘wild type distribution’, a null model for how long the longest fragment may be as a function of the number of fragments [when the virus is wild]. The red box is the ideal range for reverse genetic systems used to make infectious clones.
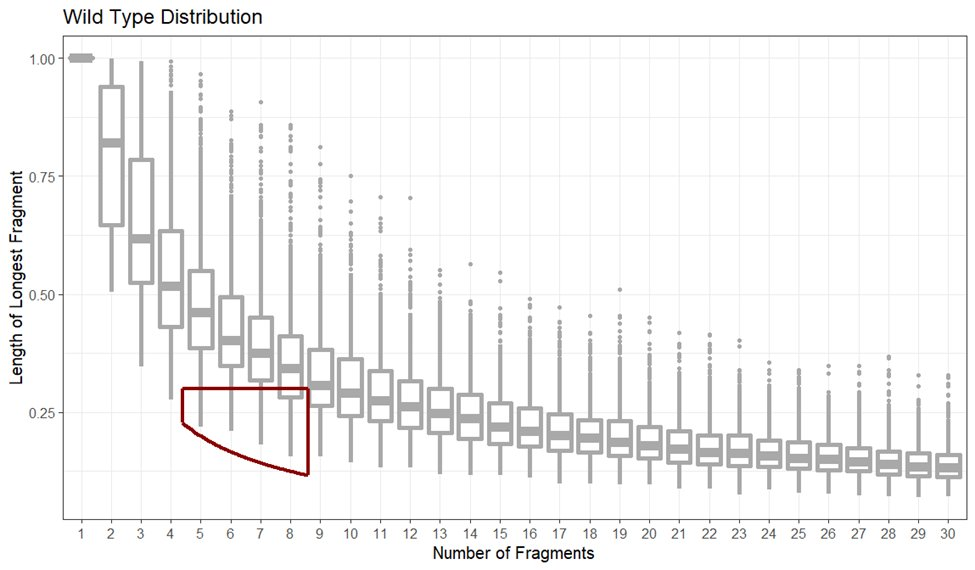
CoVs engineered to be infectious clones will move from having restriction maps falling within the wild type distribution to being outliers under the wild-type distribution, falling within the lab-ideal range of fragment number and low longest-fragment-length.
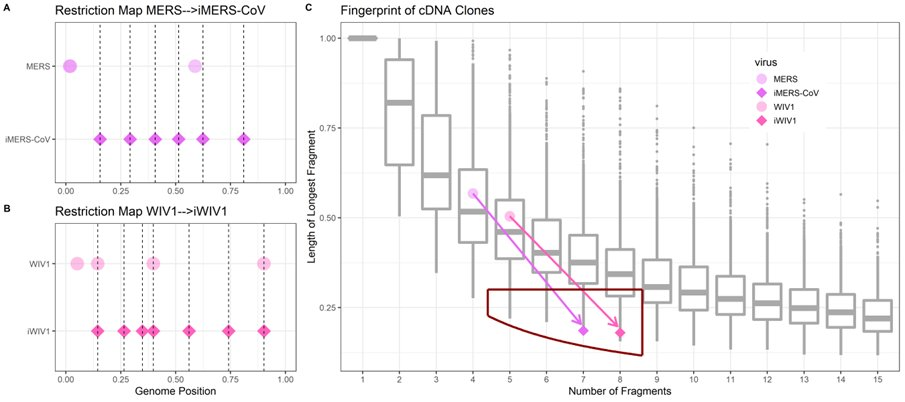
Having found this fingerprint, we examine specific cutting and pasting sites in the SARS-CoV-2 genome (BsaI/BsmBI).
BsaI and BsmBI are very popular enzymes for this kind of in vitro assembly They also have many conserved sites in CoVs. Very useful for making chimeras.
The SARS-CoV-2 BsaI/BsmBI restriction map falls neatly within the ideal range for a reverse genetic system. It is an anomaly (bottom 1%) amongst wild type CoVs. It is a midpoint amongst engineered CoVs.

Digesting CoVs with only type IIS enzymes that could be used for assembly, SARS-CoV-2 is an even greater outlier. It’s in the bottom 1% max-fragment-length for all restriction enzymes. It’s the single largest outlier (<0.07%) of 1,491 type IIS digestions.
We then tested the lab-assembly hypothesis. If SARS2 has a synthetic origin via golden gate assembly, several other criteria must be met. For example: all sticky ends must be unique, non-palindromic and contain at least one A/T. SARS2 passed this test (60% chance of this).
The mutations separating SARS-CoV-2 BsaI/BsmBI sites from its close relatives must all be silent mutations. All 14 mutations in BsaI/BsmBI sites are silent. 84% of mutations in SARS2 and close relatives are silent, so 9% chance all 14 distinct mutations will be silent.
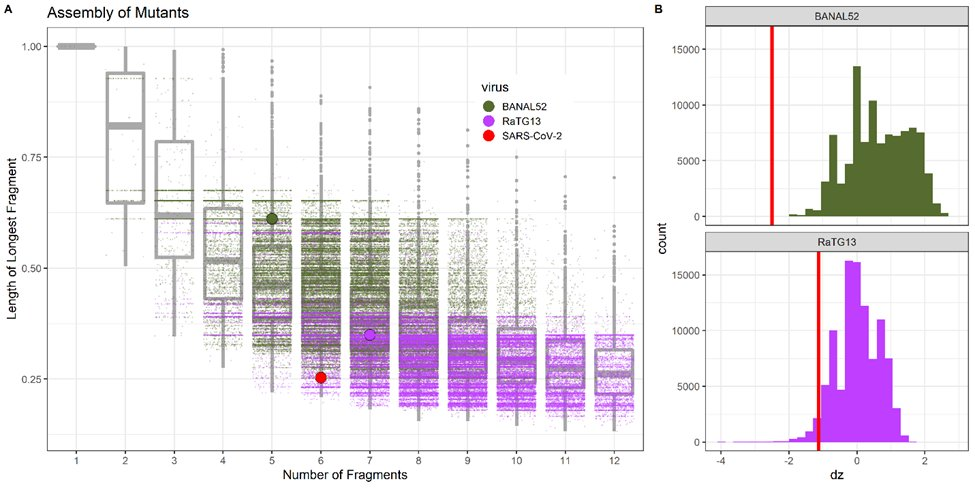
There’s a significantly higher concentration of silent mutations per nucleotide within BsaI/BsmBI recognition sequences than in the rest of the genome. P=0.004 for BANAL52-SARS2 P=9e-8 for RaTG13-SARS2.
Such an idealised reverse genetic system is unlikely to evolve by chance from the close relatives of SARS-CoV-2. There’s a 1% chance of random RaTG13 mutants having as great or greater z-score and 0.1% chance for BANAL52.
Testing this from multiple angles, we could not reject the hypothesis that SARS-CoV-2 has a synthetic origin. Each test also decreased the odds of SARS-CoV-2 having a natural origin. The BsaI/BsmBI fingerprint of SARS-CoV-2 indicates synthetic origin of SARS-CoV-2.
Please read our manuscript for our careful language and limitations. These are important. For example, our results are independent of the Furin Cleavage Site (FCS). While the RBD is docked in fragment 5, we shine no light on the origin of the FCS.
Our research does not identify the lab. We hypothesise this restriction map would enable construction of chimeric viruses, much like the recent controversial work done in Boston (but with a different method for in vitro assembly).
Our theory of a synthetic origin of SARS-CoV-2 can and should be tested. Further tests may reject our theory. We welcome these tests. Our code is available on GitHub and we point to future research that can reject our hypothesis or refine our understanding of this issue.
Making chimeric viruses in vitro carries risks. We encourage transparency from researchers studying CoVs in Wuhan. We strongly encourage global coordination on biosafety.
We encourage open, civil and compassionate discourse on this important topic. This pre-print was not rushed. It was reviewed by many colleagues, truly world experts. We thank them all immensely for their feedback. This has been an incredible project. Yet, for obvious reasons, this is the saddest paper I’ve ever written.
Stop Press: The paper has been heavily criticised by multiple specialists on Twitter, who have highlighted what they deem to be fatal flaws in it. Prof. Balloux has unexpectedly deleted his Twitter account, apparently in light of the criticisms of the paper, though he has not confirmed this. Dr. John Ross has (somewhat snidely) tweeted: “In case you missed it, U.K. Covid minimiser Francois Balloux has deleted his account after amplifying a spectacularly wrong preprint claiming to have proven that SARS-CoV-2 was synthetic (refuted below).”
Dr. Ross’s Twitter thread and the replies to it provide links to a number of criticisms of the paper.
Dr. Washburne has responded here and here defending his paper.
Matt Ridley has praised the response for being reasoned and without invective: “This thread is a good response to a reasonable criticism. Not an insult in sight. Science as it should be done.”









To join in with the discussion please make a donation to The Daily Sceptic.
Profanity and abuse will be removed and may lead to a permanent ban.
“Professor Francois Balloux has given the study his imprimatur” I stopped reading righ there.
Didn’t we already know that Moderna patented the s-protein variants in 2016?
https://pubchem.ncbi.nlm.nih.gov/patent/US-9587003-B2
No wonder they are suing Pfizer for stealing their idea for the jabs.
Go on admit it. You did not understand a single word of this gobbledygook.
And this is the problem with Covid 19, the scientists will always invite you to fight on their own turf and, funnily enough, they always end up winning. That’s because there are a handful of scientists on earth that can use this type of genetic gobbledygook to prove or disprove anything they want because no one else knows what the hell they’re are talking about.
Despite all that, they still cannot answer the simple questions. This person may have found proof that this is not a bioweapon but that was clear to anyone with half a brain at the very beginning of this ‘pandemic’. Simply put, if this was a bioweapon leaked from a lab it was the worst designed bioweapon ever made as it only managed to kill elderly people at an age ABOVE the average age of natural death!
Yes, it was obvious that the idea that this was a bioweapon was simply ludicrous and you didn’t need a degree in molecular genetics to understand that.
As the saying goes “If you can’t explain it simply, you don’t understand it well enough.”
Remember that and all these scientific word salads start to make make sense.
I think the idea behind the bio-weapon theory is that they were doing gain of function research to make viruses more infectious, not necessarily more deadly. I think it might be an apocryphal tale, because I only heard it post-Covid, but the Spanish flu is said to have been the straw that broke the camels back that led to the Germans surrendering WWI. I understand the one of the purposes of gain of function is to produce a bio-weapon that is virulent, but not deadly enough to annihilate mankind.
The spike protein is a toxic bioweapon engineered to have greater affinity to certain racial ACE2 receptor genotypes. I have this information on good authority from a military MD acquaintance. Anyone who is not a K26R genotype is susceptible, with white Europeans being specifically targeted.
ACE2 coding variants in different populations and their potential impact on SARS-CoV-2 binding affinity
No wonder East Asians got the least deaths despite numerous cases. That, and they probably already had a proto-variant in 2019 which gave them some prior immunity.
Called SARS from 2003 in fact, that’s why.
I always heard the only reason the Germans surrendered was because America joined in the war. So that means that America could have stayed out of the war and the outcome for Europe would have been more or less the same. But then again, the “Spanish” Flu really originated in Fort Funston, Kansas, and was spread to Europe by American troops where it incubated in the trenches and became nastier. So had we stayed out of the war, the pandemic may not have even happened at all.
Doubletalk, doublespeak, gobbledygook, blinding with science, word salad, Occam’s Butterknife (opposite of Occam’s Razor) — whatever you call it, it is all to obfuscate what really happened and make it look far more benign than it really is.
This is a clear example of two distinct and separate subjective realities that people inhabit in the same objective world.
The first believe that there was a virus. That it has been sequenced and this has been done so well that we can inspect the code and discover anomalies that look like a synthetic signature.
The second group believe there is no virus. They principally believe this because the virus has never been isolated, and therefore the DNA sequence may be polluted by other DNA in the sample. The virus can be explained as rebranded seasonal flu, coupled with a global propaganda campaign and some hooky “the science” to grease the wheels.
Both beliefs believe they are true.
If the first is true then I think it places a giant question mark over the vaccine and its purpose. Certainly none of the claims made about its safety and effectivity have proven to be true. Unless of course one is to believe the completely unprovable claim over the millions of lives it is professed to have saved.
I’m so glad that all these highly educated, highly qualified, scientist’s studies are coming to exactly the same conclusion that many of us came to more than two years ago using that rarest of abilities – common sense.
Very interesting. One thing I’ve heard said is that artificial or genetically modified organisms have a tendency over time and generations to revert to their natural form, almost like nature resetting itself and erasing the aberration. I’ve been wondering whether this is why we see the coronavirus gradually behaving more like a regular respiratory virus, i.e. less virulence and less transmissibility?
On the other hand, like many others, I don’t believe that the virus itself has done anywhere near as much harm as the so-called responses imposed to deal with it.
Indeed.
Stands to reason, evolutionary pressures are very ancient.
Having looked at the original paper I thought the summary here was pretty clear and the last three paras, indicating a willingness to be proved wrong, the recognition of risk and his very last line, show a degree of humiliation – nay, even proper ‘science’ – that is sadly lacking in the consensus crowd. But the most chilling line in the paper that clinched it for me is this: “Additionally, our wild type distribution drew on a wide range of 70 non-SARS-COV-2 genomes and 214 restriction enzymes for sale at New England Biosciences,….” [my emphasis] indicating just how far they’ve gone down this particular rabbit hole already.
.
The smoking gun right here. But it was somehow not a bioweapon or GOF research. Move along, nothing to see here folks….
Remember that the Wuhan Institute of Virology did not apply strict safety precautions to the SARS GoF research, essentially this meant use gloves but leave your lab coat flapping open and don’t have any additional pressure-reduced labs and airlocks etc.
They didn’t appear to think it was particularly dangerous work, and when their work inevitably escaped into the external world the effects were not clearly understood. The real problem was the labelled spike proteins (sequences not seen in nature used to keep track of the thing in the lab) but the furin cleavage site added caused much higher transmissivity and made the spike protein a weapon in itself. I suspect this last was an accident. Add all these things together and you get greedy people accidentally releasing something they didn’t recognise as nasty by mistake and then other greedy people grabbing hold of the lab-created and known spike protein to use as a basis for their experimental and very dangerous mRNA vaccines.
Just like aircraft crashes it’s always a sequence of things going wrong, rarely just one thing.
What we know for sure is that the “theory” that the virus spread from some “wet market” in Wuhan in December 2019 is preposterous. This virus didn’t even “escape” from a lab in November, which was the big story at one time after the Wall Street Journal published a scoop that U.S. intelligence had learned of three lab workers who became ill and had to be hospitalized in “November” 2019. As I’ve posted too many times to count, copious evidence exists that the virus was spreading at least by October 2019, which is when the World Military Games were held in Wuhan (and many athletes became sick from something).
RFK, Jr. in the new documentary about his book mentions two times that Chinese military officials took over the operation of the WIV lab on September 12th 2019 and seemed to scrub a lot of evidence. U.S. intelligence satellite photos reportedly show many hospitals in China that featured parking lots overflowing with cars … in September.
If the virus that caused those athletes to get sick in October was the novel coronavirus, this virus must have been spreading for many weeks before the start of those Games. Kennedy says the spread began in September if not August 2019. He also strongly suggest Chinese officials knew this. My conjecture is that at least some American officials probably knew this as well.
This much-earlier virus “birth date” would have allowed the virus plenty of time to spread to most countries in the world, including America by October or early November 2019.
If this hypothesis/theory is true, one can’t help but wonder what was all that footage about showing Chinese falling dead in the streets in late December 2019?
Anyway, to find the origins of the escape or birth of this virus, virus sleuths need to go back many months earlier. Why they refuse to do this is another interesting question.
If researchers/investigators were sincere about finding the true story of the virus’s origins, they would keep working backwards in an effort to find the real “case zero.” They would interview anyone and everyone who might have been infected before the previous first-known case. They would test these people for antibodies, interview their close contacts, check any medical records they might have, etc. But none of this has happened. My take-away: This is NOT a sincere investigation into the real origins of this virus.
Someone asked me at a dinner party last year. “So Hoppy, wet market or lab leak?”
Hoppy “Oh, lab leak.”
Interrogator, sitting back to be entertained “Oh, why?”
Hoppy “Body language of the world’s governments. They’ve never acted like this before. Shutting everything down, going on about how it’s a totally new virus, it might not evolve like normal viruses to be less virulent etc. They clearly thought it was an escaped bioweapon.”
(Silence).
Historically science has evolved from alchemy – trying to produce gold for gullible rulers out of lead. I have a feeling that the PTB believe in the virology and bio-weapon scenario and that the Chinese have something far deadlier hidden up their sleeves whereas the Emperor’s New Clothes are missing in plain sight.